My lab is interested in the mechanisms that control developmental gene regulation and the crucial transformations that enabled the evolution of complex multicellularity. Our research combines molecular, developmental, genomic and evolutionary approaches. We currently study genome-wide quantitative DNA accessibility and transcription at regulatory elements during sea urchin development. This is helping to map the precise location and activity of proximal and distal gene regulatory elements at the same time that we identify specific mechanisms that set developmental potency in early embryos and stem cells. We are also performing genomic and developmental reprogramming experiments to functionally test the relationships among DNA, nucleosomes and transcription factors that modulate transcriptional potency. In collaboration with researchers at Cornell University, Ithaca, and the Institute of Evolutionary Biology in Barcelona, we are further exploring which regulatory mechanisms facilitated the evolution of metazoan developmental gene regulatory networks.
Degrees
BS. Universitat de Barcelona
Ph.D. CID/CSIC and Universitat de Barcelona
Postdoctoral research at Caltech
Full List of publications at:
https://www.ncbi.nlm.nih.gov/myncbi/cesar.arenas-mena.2/bibliography/public/
Selected publications
Arenas-Mena, Cesar, and Serhat Akin. 2023. “Widespread Priming of Transcriptional Regulatory Elements by Incipient Accessibility or RNA Polymerase II Pause in Early Embryos of the Sea Urchin Strongylocentrotus Purpuratus.” Genetics, August, 2023, iyad145. https://doi.org/10.1093/genetics/iyad145
Alexandra G. Chivu, Abderhman Abuhashem, Gilad Barshad, Edward J. Rice, Michelle M. Leger, Albert C. Vill, Wilfred Wong, Rebecca Brady, Jeramiah J. Smith, Athula H. Wikramanayake, César Arenas-Mena, Ilana L. Brito, Iñaki Ruiz-Trillo, Anna-Katerina Hadjantonakis, John T. Lis, James J. Lewis, Charles G. Danko. (Submitted). Evolution of promoter-proximal pausing enabled a new layer of transcription control. bioRxiv 2023.02.19.529146; doi: https://doi.org/10.1101/2023.02.19.529146
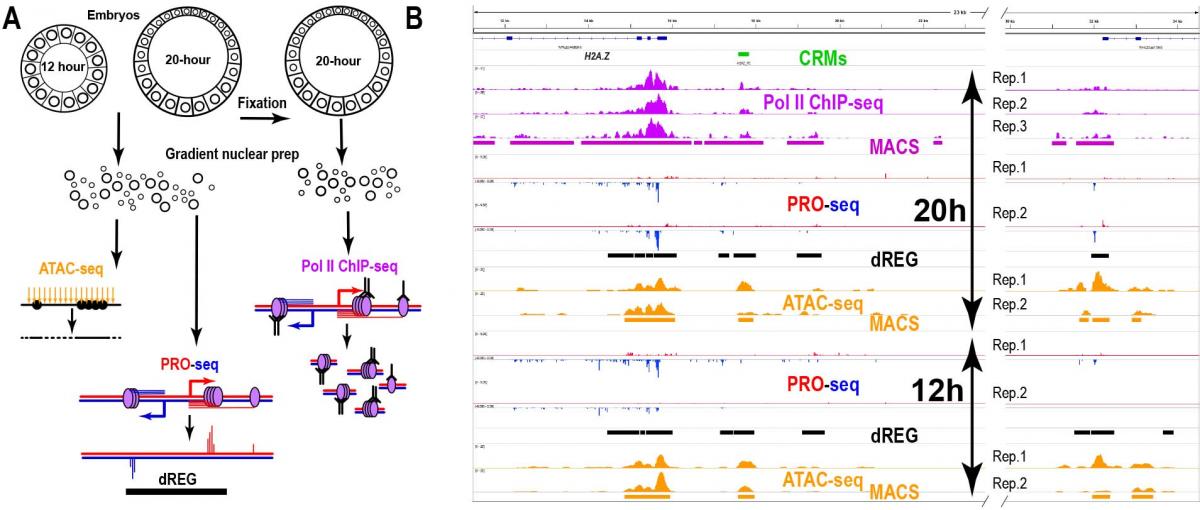
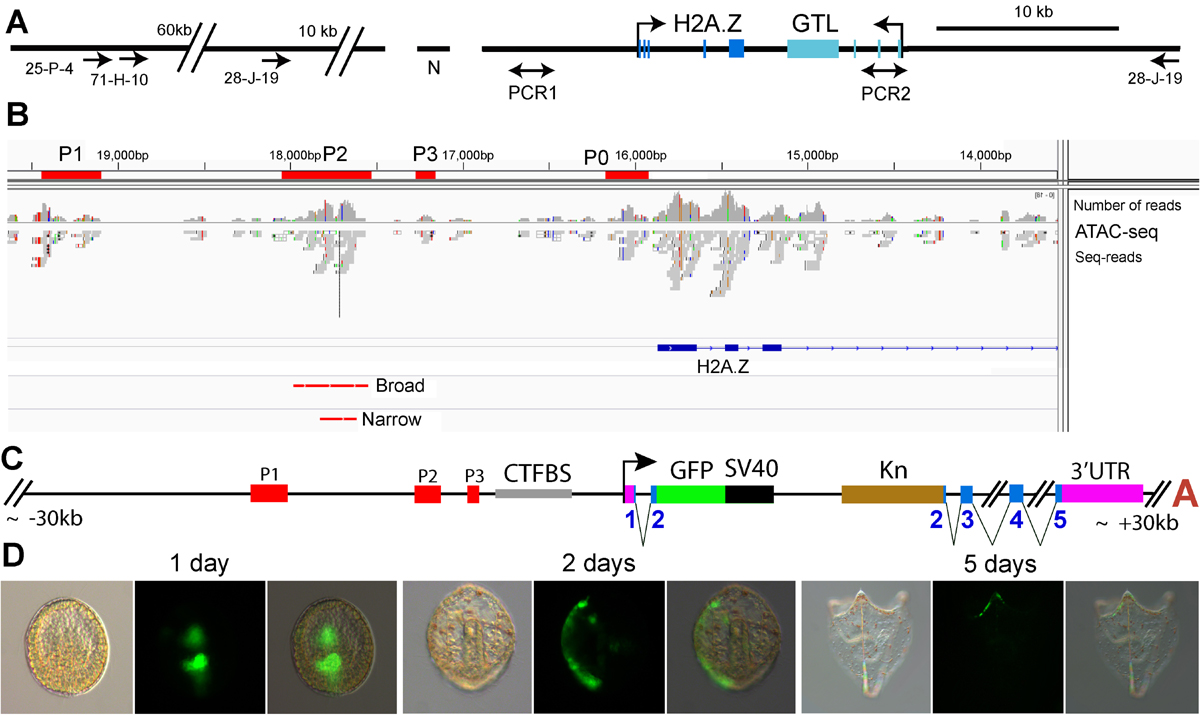
Arenas-Mena, C., Miljovska, S., Rice, E. J., Gurges, J., Shashikant, T., Wang, Z., Ercan, S., & Danko, C. G. (2021). Identification and prediction of developmental enhancers in sea urchin embryos. BMC Genomics, 22(1), 751. https://bmcgenomics.biomedcentral.com/articles/10.1186/s12864-021-07936-0
Arenas-Mena, C. (2017). The origins of developmental gene regulation. Evolution & Development, 19(2), 96–107. https://doi.org/10.1111/ede.12217
Hajdu, M., Calle, J., Puno, A., Haruna, A., & Arenas-Mena, C. (2016). Transcriptional and post-transcriptional regulation of histone variant H2A.Z during sea urchin development. Development, Growth & Differentiation, 9(3), 231–243. https://doi.org/10.1111/dgd.12329
Wong, K. S.-Y., & Arenas-Mena, C. (2016). Expression of GATA and POU transcription factors during the development of the planktotrophic trochophore of the polychaete serpulid Hydroides elegans. Evolution & Development, 254–266. https://doi.org/10.1111/ede.12196
Arenas-Mena, C., & Coffman, J. A. (2015). Developmental control of transcriptional and proliferative potency during the evolutionary emergence of animals. Developmental Dynamics, 244(10), 11093–11201. https://doi.org/10.1002/dvdy.24305
Arenas-Mena, C., & Li, A. (2014). Development of a feeding trochophore in the polychaete Hydroides elegans. The International Journal of Developmental Biology, 58, 575–583. https://doi.org/10.1387/ijdb.140100ca
Arenas-Mena, C. (2010). Indirect development, transdifferentiation and the macroregulatory evolution of metazoans. Philosophical Transactions of the Royal Society B: Biological Sciences, 365(1540), 653–669. https://doi.org/10.1098/rstb.2009.0253
Arenas-Mena, C. (2007). Developmental transcriptional-competence model for a histone variant and a unicellular origin scenario for transcriptional-multipotency mechanisms. Evolution & Development, 9(3), 208–211. https://doi.org/10.1111/j.1525-142X.2007.00156.x
Arenas-Mena, C., Wong, K. S.-Y., & Arandi-Foroshani, N. R. (2007). Histone H2A. Z expression in two indirectly developing marine invertebrates correlates with undifferentiated and multipotent cells. Evolution & Development, 9(3), 231–243. https://doi.org/10.1111/j.1525-142X.2007.00155.x
Davidson, E. H., Rast, J. P., Oliveri, P., Ransick, A., Calestani, C., Yuh, C.-H., Minokawa, T., Amore, G., Hinman, V., Arenas-Mena, C., Brown, C. T., Livi, C. B., Revilla, R., Rust, A. G., Pan, Z., Schilstra, M. J., Clarke, P. J. C., Arnone, M. I., Rowen, L., … Bolouri, H. (2002). A Genomic Regulatory Network for Development. Science, 295(5560), 1669–1678. https://doi.org/10.1126/science.1069883
Research
• Genome-wide characterization of transcriptional regulatory elements
We are performing genome-wide analysis of sea urchin transcriptional regulatory elements during development. We use ATAC-seq, PRO-seq to identify and functionally characterize the enhancers and promoters that control sea urchin development (Arenas-Mena and Akin, 2023; Arenas-Mena et at., 2021). The research questions relate to transcriptional networks and the unicellular origins of developmental gene regulation (Arenas-Mena, 2017). In particular, we are interested in the unicellular precursors of distal enhancers.
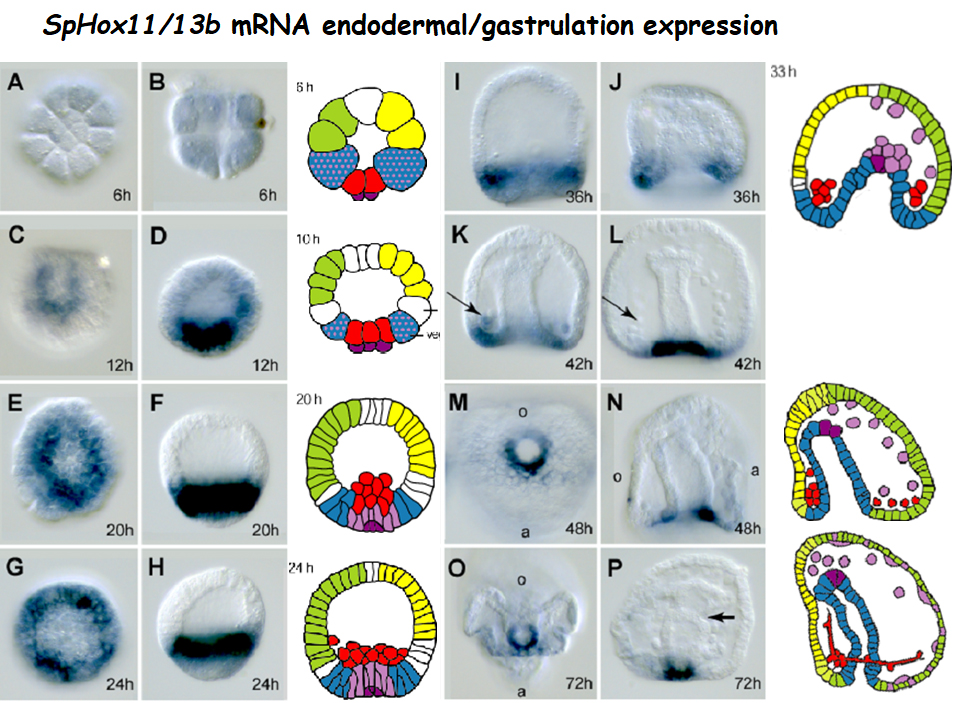
• Developmental control of transcriptional and proliferative potency
My original hypothesis of a histone variant H2A.Z role in developmental potency (Arenas-Mena, 2007; Arenas-Mena et al., 2007) has been validated experimentally (Arenas-Mena and Coffman, 2015). H2A.Z is associated with transcriptional regulatory DNA, where it promotes an open chromatin state accessible to sequence-specific transcription factors. We study of the cis-regulatory machinery that controls the developmental expression of H2A.Z (Hajdu et al., 2016). In addition, we have elaborated a method of developmental reprogramming Doxycycline-inducible expression , and we are testing how H2A.Z maintains transcriptional potency during development.
• A new polychaete system relevant to bilaterian body plan evolution
We have spearheaded the implementation of methods and resources in the annelid Hydroides elegans, an indirectly developing polychaete with feeding trochophore relevant to bilaterian body plan evolution (Arenas-Mena and Li, 2014). We are currently undertaking transgenic and genomics approaches in this new model.
Current Lab Members
Serhat Akin, PhD student, MCD program.
Syeda Batool. Master's Student
Former Lab Members
PhD
Mihai Hajdu
Undergraduate
Basimah Sibawe
Maria Graham
Jasmine Calle
Andrea Puno
Aminat Haruna
Justin Gurges
Krystal Baird
Winnie Darius
Navid Arandi
Tia Leung
Fedan Avrumova
Crystal Lucas
Merlin Raj
Jose Alvarenga
Leila Lager
Alfonso Clemente
Andrew Fischler
Ashley Woods
Joseph Deas
Kasey Mobley
Moriel Khaykin
Rossana Cruciata
Charisse White
Dimitri Mazidis
Jacob Maddela
Lisa Lamanna
Mark Montano
Monique Brewton
Natalie Birkin
Ojeh Agnes
Master's
Zihe Wang
Kimberly Suk-Ying Wong
Ava Li,
Shona Shonghai
Nayomi Fernando
Dasari Srividya
Triveni Golagani
Organisms
Strongylocentrotus purpuratus
Research in sea urchins has lead the experimental characterization of developmental gene regulatory networks thanks to their experimental and biological. Genomic resouces are available at EchinoBase. Cover from (Arenas-Mena et al., 2000).
![]()
Arenas-Mena et. al, 2000

Arenas-Mena et. al, 2006
Hydroides elegans
We have lead efforts to develop a polychaete model system with spiral cleavage and feeding trochophore that has great evolutionary and developmental relevance (Arenas-Mena and Li, 2014). Cover from (Arenas-Mena, 2013).

Arenas-Mena, 2013
Funding and Collaborators
Teaching
CUNY Graduate Center
Developmental Biology
Biochemistry
CSI
Genomics Laboratory
Developmental Biology
Gene Regulatory Systems
Cell Biology
Evolution of Development
Invertebrate zoology
General Biology

Contact Information
Research opportunities
I am currently recruiting Ph.D. students from the graduate programs in
- Molecular, Cellular and Developmental Biology
- Biochemistry
- Ecology, Evolutionary Biology and Behaviour
I am also accepting highly motivated Master's and undergraduate students.