Our lab is interested in the mechanisms that control developmental gene regulation, transcriptional multipotency and the crucial transformations that enabled the evolution of complex multicellularity. Our research combines molecular, biochemical, developmental, evolutionary and genomic approaches. We currently study genome-wide quantitative DNA accessibility and transcription at regulatory elements during sea urchin development. This is helping to map the precise location and activity of proximal and distal gene regulatory elements at the same time that we identify specific transcriptional regulatory mechanisms that set developmental potency in early embryos and stem cells. We are performing genomic and developmental reprogramming experiments to functionally test the relationships among DNA, nucleosomes and transcription factors that modulate transcriptional potency. We are currently elaborating new methods for the high-throughput analysis of transcriptional regulatory elements.
Degrees
BS. Universitat de Barcelona
Ph.D. CID/CSIC and Universitat de Barcelona
Postdoctoral research at Caltech
Full List of publications at:
https://www.ncbi.nlm.nih.gov/myncbi/cesar.arenas-mena.2/bibliography/public/
Selected publications
Chivu, A. G., Basso, B. A., Abuhashem, A., Leger, M. M., Barshad, G., Rice, E. J., Vill, A. C., Wong, W., Chou, S.-P., Chovatiya, G., Brady, R., Smith, J. J., Wikramanayake, A. H., Arenas-Mena, C., Brito, I. L., Ruiz-Trillo, I., Hadjantonakis, A.-K., Lis, J. T., Lewis, J. J., & Danko, C. G. (2025). Evolution of promoter-proximal pausing enabled a new layer of transcription control. Nature Structural & Molecular Biology. https://doi.org/10.1038/s41594-025-01718-y
Arenas-Mena, Cesar, and Serhat Akin. 2023. “Widespread Priming of Transcriptional Regulatory Elements by Incipient Accessibility or RNA Polymerase II Pause in Early Embryos of the Sea Urchin Strongylocentrotus Purpuratus.” Genetics, August, 2023, iyad145. https://doi.org/10.1093/genetics/iyad145
Arenas-Mena, C., Miljovska, S., Rice, E. J., Gurges, J., Shashikant, T., Wang, Z., Ercan, S., & Danko, C. G. (2021). Identification and prediction of developmental enhancers in sea urchin embryos. BMC Genomics, 22(1), 751. https://bmcgenomics.biomedcentral.com/articles/10.1186/s12864-021-07936-0
Arenas-Mena, C. (2017). The origins of developmental gene regulation. Evolution & Development, 19(2), 96–107. https://doi.org/10.1111/ede.12217
Hajdu, M., Calle, J., Puno, A., Haruna, A., & Arenas-Mena, C. (2016). Transcriptional and post-transcriptional regulation of histone variant H2A.Z during sea urchin development. Development, Growth & Differentiation, 9(3), 231–243. https://doi.org/10.1111/dgd.12329
Wong, K. S.-Y., & Arenas-Mena, C. (2016). Expression of GATA and POU transcription factors during the development of the planktotrophic trochophore of the polychaete serpulid Hydroides elegans. Evolution & Development, 254–266. https://doi.org/10.1111/ede.12196
Arenas-Mena, C., & Coffman, J. A. (2015). Developmental control of transcriptional and proliferative potency during the evolutionary emergence of animals. Developmental Dynamics, 244(10), 11093–11201. https://doi.org/10.1002/dvdy.24305
Arenas-Mena, C., & Li, A. (2014). Development of a feeding trochophore in the polychaete Hydroides elegans. The International Journal of Developmental Biology, 58, 575–583. https://doi.org/10.1387/ijdb.140100ca
Arenas-Mena, C. (2010). Indirect development, transdifferentiation and the macroregulatory evolution of metazoans. Philosophical Transactions of the Royal Society B: Biological Sciences, 365(1540), 653–669. https://doi.org/10.1098/rstb.2009.0253
Arenas-Mena, C. (2007). Developmental transcriptional-competence model for a histone variant and a unicellular origin scenario for transcriptional-multipotency mechanisms. Evolution & Development, 9(3), 208–211. https://doi.org/10.1111/j.1525-142X.2007.00156.x
Arenas-Mena, C., Wong, K. S.-Y., & Arandi-Foroshani, N. R. (2007). Histone H2A. Z expression in two indirectly developing marine invertebrates correlates with undifferentiated and multipotent cells. Evolution & Development, 9(3), 231–243. https://doi.org/10.1111/j.1525-142X.2007.00155.x
Davidson, E. H., Rast, J. P., Oliveri, P., Ransick, A., Calestani, C., Yuh, C.-H., Minokawa, T., Amore, G., Hinman, V., Arenas-Mena, C., Brown, C. T., Livi, C. B., Revilla, R., Rust, A. G., Pan, Z., Schilstra, M. J., Clarke, P. J. C., Arnone, M. I., Rowen, L., … Bolouri, H. (2002). A Genomic Regulatory Network for Development. Science, 295(5560), 1669–1678. https://doi.org/10.1126/science.1069883
Research
• Transcriptional Potency in Early Embryos. NIH funded. One PhD position available.
This project seeks to identify and test mechanisms that establish the transcriptional regulatory potency of early embryos. The relationships among DNA accessibility, nucleosome lability, transcription, histone variants and DNA sequences interacting with nucleosomes and transcription factors in early embryos and differentiated cell types are identified with genomic methods and experimentally tested. Our prior results (Arenas-Mena and Akin, 2023) suggest that regulatory potential may be primed at some transcriptional regulatory elements (TREs, inclusive of promoters and enhancers/silencers) by DNA sequences promoting weak histone interactions. Such sequence-based priming may synergize with the embryonic abundance of histone variants known to promote TRE accessibility. In particular, H2A.Z/H3.3 histone variant nucleosomes associate with low occupancy TREs. Furthermore, H2A.Z mRNA expression is high in blastula embryos and multipotent adult cell precursors of indirectly developing sea urchins and polychaetes but low in differentiated cells (Arenas-Mena et al., 2007), therefore supporting a general H2A.Z transcriptional multipotency role (Arenas-Mena, 2007).
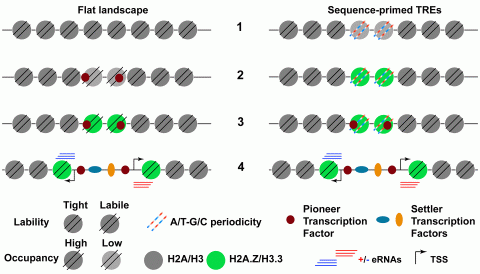
Hypothesis. Generic sequence of events, 1 to 4, leading to TRE activation in flat accessibility landscape TREs (left) and sequence-primed TREs (right). Left, early during development, TREs are “hidden” in a generally flat chromatin accessibility landscape, 1; until unlocked by zygotic pioneer transcription factors that recruit chromatin enzymatic activities, which loosen DNA-histone interactions, lower nucleosome occupancy, 2, and replace canonical replication histones with H2A.Z and H3.3 histone variants; 3, this further increases DNA accessibility and facilitates settler transcription factor binding that eventually triggers TRE transcriptional activation, 4. Right, in sequence-primed TREs, regulatory elements inherently have incipient accessibility thanks to A/T-G/C sequence periodicities that weaken DNA interactions with canonical histones H2A and H3 and cause labile nucleosomes, 1, that are replaced by highly abundant histone variants H3.3 and H2A.Z during embryogenesis, 2, and facilitate pioneer transcription factor activity, 3, eventually leading to TRE activation, 4, or stable silencing (not shown). Alternative chromatin modifications, other than remodeling or histone variants, and/or sequence of events (2 to 3) would fit both models, which are non-exclusive, and are expected to be found at TREs with distinct biological functions. eRNAs, enhancer RNAs (or upstream and coding RNAs in promoters); TSS, transcription start site.
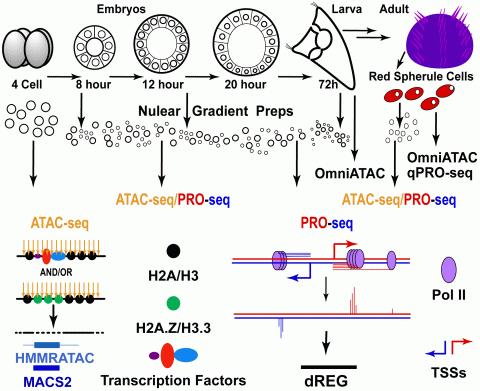
PRO-seq (Precision Run-On Sequencing, a method to detect actively transcribing TREs) followed by dREG (Discriminative Regulatory Element Detection, a vector machine TRE prediction tool) will identify transcriptionally active TREs in early embryos and differentiated adult cell types (Arenas-Mena et at., 2021). OmniATAC experiments will test if in early embryo and differentiated cell TREs have incipient DNA accessibility. Quantitative micrococcal nuclease and sequencing (qMNase-seq) will test if TREs with incipient DNA accessibility in early embryos are occupied by labile nucleosomes. H2A.Z and H3.3 CUT&RUN (Cleavage Under Targets & Release Using Nuclease) will test the prevalence of these histone variants at labile nucleosomes. The new data sets will synergize with existing ATAC-seq and PRO-seq characterizations during sea urchin embryogenesis and differentiation. Comparison among genomic profiles will test if transcriptionally disengaged but accessible TREs in early embryos associate with labile nucleosomes, H2A.Z, H3.3, A/T-GC sequence periodicities and/or pioneeer transcription factor binding sites (TFBSs). Similar experiments in terminally differentiated cell types with low H2A.Z expression will test if incipient accessibility is an inherent property of TREs entirely based on A/T-GC sequence periodicities. Accessibility sequences setting incipient TRE accessibility during embryogenesis will be experimentally tested by DNase-I-seq of zygotically microinjected TREs harboring mutations that alter A/T-GC sequence periodicities or pioneer TFBSs. The project will be integrated with parallel efforts to implement “research in the classroom” in existing genomic courses that will recruit, train and motivate diverse undergraduate and graduate students at CUNY. The understanding of basic transcriptional multipotency mechanisms in sea urchin embryos is relevant to understand the evolution of developmental gene regulation (Arenas-Mena, 2017, Arenas-Mena, 2010) and to advance future therapeutics of tissue repair and regeneration.
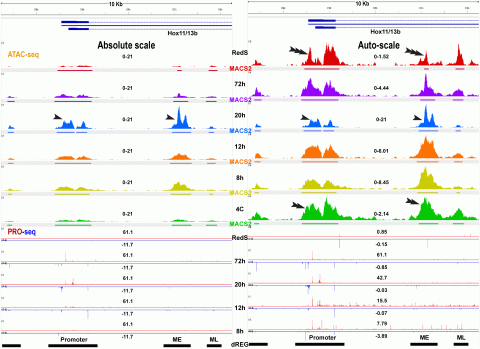
Outline of ATAC- and PRO-seq experiments (Arenas-Mena et at., 2021). ATAC-seq detects DNA accessibility and PRO-seq the last nucleotide incorporated by transcribing or paused RNA polymerases. ATAC-seq peak calls are made by MACS2 and HMMRATAC, with thick lines indicating regions of sub-nucleosomal read length and thin lines flanking nucleosomes. TRE predictions from PRO-seq data are made by dREG (Discriminative Regulatory Element Detection), a machine learning method.
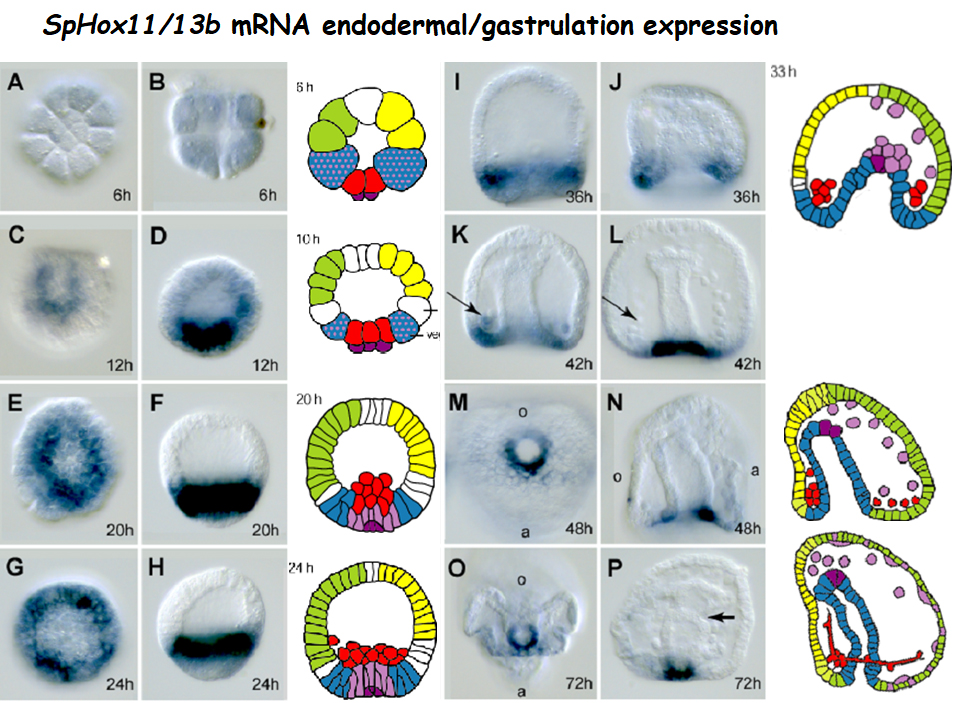
Our results (Arenas-Mena et at., 2021) revealed incipient TRE accessibility in 4 cell embryos, which may be due to transcription factor binding and/or labile nucleosome occupancy. Example of ATAC- and PRO-seq genomic profiles during embryonic development and differentiation at the Hox11/13b locus. Genomic profiles in reads per million. The PRO-seq run-on transcripts/base is shown for each base in the plus (red, top) and minus (blue, bottom) strands, with PRO-seq peaks marking the transcriptional pause sites of TSSs. The left panel (Absolute scale) shows ATAC-seq profiles set at the maximum scale range of the 20 hour sample in reads per million (0-21), and the right panel (Auto-scale) with each range set to the maximum value of each track. For PRO-seq the absolute scale is independently set for the plus and minus strands among all samples, with negative values for the minus strand. The dREG TRE predictions are the combined and merged set of the 8, 12, 20, 72 hour and red spherule cells dREG predictions. Bottom, ME and ML are enhancers whose activity was experimentally tested in reporter constructs (Cui et al., 2017). 4C, 4 cell embryos; 8-20 h embryos and 72 h larva as indicated; H2A.Z and H3.3 are histone variants; Pol II, RNA polymerase II; RedS, differentiated red spherule cells; TSSs, transcription start sites.
• Cis-regulatory analysis of Cyclin D. Cyclin D is a major control of cell proliferation and it is developmentally regulated at the transcriptional level (Arenas-Mena and Coffman, 2015). Cell proliferation largely contributes to the sophisticated shapes of metazoans and its missregulation leads to cancer. We leverage our genomic data sets and the advantages of sea urchins for transcriptional regulatory analysis to identify the TREs that control the expression of Cyclin D during embryonic and postembryonic development.
• New methods for the analysis of transcriptional regulatory elements. Our genomic data sets are combined with new methods that facilitate the analysis of transcriptional regulatory elements at single cell resolution.
• Transcriptional multipotency role of Histone variant H2A.Z. The essential role of high H2AZ levels in early embryos and multipotent adult precursors is experimentally tested.
• A new polychaete system relevant to bilaterian body plan evolution
We have spearheaded the implementation of methods and resources in the annelid Hydroides elegans, an indirectly developing polychaete with feeding trochophore relevant to bilaterian body plan evolution (Arenas-Mena and Li, 2014). We are currently undertaking transgenic and genomics approaches in this new model.
Current Lab Members
Serhat Akin, PhD student, Molecular Cell and Developmental Biology Subprogram
Pijush Sutradhar, Postdoc
Jawuneh Danmbisaa, Biology Master's Student
Marina Anastasiadis Undergraduate Student
Selma Musovski Undergraduate Student
Roqaya Hassan Undergraduate Student
Former Lab Members
PhD
Mihai Hajdu
Master's
Syeda Batool
Zihe Wang
Kimberly Suk-Ying Wong
Ava Li
Shona Shonghai
Nayomi Fernando
Dasari Srividya
Triveni Golagani
Undergraduate
Basimah Sibawe
Maria Graham
Jasmine Calle
Andrea Puno
Aminat Haruna
Justin Gurges
Krystal Baird
Winnie Darius
Navid Arandi
Tia Leung
Fedan Avrumova
Crystal Lucas
Merlin Raj
Jose Alvarenga
Leila Lager
Alfonso Clemente
Andrew Fischler
Ashley Woods
Joseph Deas
Kasey Mobley
Moriel Khaykin
Rossana Cruciata
Charisse White
Dimitri Mazidis
Jacob Maddela
Lisa Lamanna
Mark Montano
Monique Brewton
Natalie Birkin
Ojeh Agnes
Organisms
Strongylocentrotus purpuratus
Research in sea urchins has lead the experimental characterization of developmental gene regulatory networks thanks to their experimental and biological. Genomic resouces are available at EchinoBase. Cover from (Arenas-Mena et al., 2000).
![]()
Arenas-Mena et. al, 2000

Arenas-Mena et. al, 2006
Hydroides elegans
We have lead efforts to develop a polychaete model system with spiral cleavage and feeding trochophore that has great evolutionary and developmental relevance (Arenas-Mena and Li, 2014). Cover from (Arenas-Mena, 2013).

Arenas-Mena, 2013
Funding and Collaborators
CURRENTLY FUNDED PROJECTS

NIH Award 1R15HD115134 . NATIONAL INSTITUTE OF CHILD HEALTH & HUMAN DEVELOPMENT.
PRIOR FUNDING
NASA Exobiology

PSC-CUNY

NIH
COLLABORATORS:
Teaching
CUNY Graduate Center
Developmental Biology
Biochemistry
CSI
Genomics Laboratory
Developmental Biology
Gene Regulatory Systems
Cell Biology
Evolution of Development
Invertebrate zoology
General Biology

Contact Information
Research opportunities
There is 1 PhD position available for students in the GC programs:
I would like to mentor students interested in evolutionary projects:
I am also recruiting highly motivated Master's and undergraduate students.